Merger trees
We provide merger trees describing the build-up of haloes. These are built using the LHaloTree code and saved in HDF5 files. These are identical to the merger trees provided by the IllustrisTNG simulations. A thorough description of the data format is provided at the end of this page.
The following tables (click to open) describe the fields. First, the Header group:
| Header datasets ▾ |
|---|
| Attribute | Dimensions | Units | Description |
|---|---|---|---|
| Redshifts | Nsnap | - | List of the redshift of the snapshots used to create the merger tree. |
| TotNsubhalos | Nsnap | - | Equal to the number of Subfind/Subhalo groups in the group catalog, for each snapshot used to create this merger tree. |
| TreeNHalos | Nhalos | - | The number of entrues in each TreeX group in this file. |
| Header attributes ▾ |
|---|
| Attribute | Dimensions | Units | Description |
|---|---|---|---|
| FirstSnapshotNr | 1 | - | First snapshot number used to make these merger trees (should be 0). |
| LastSnapshotNr | 1 | - | Last snapshot number used to make these merger trees (varies for different runs). |
| SnapSkipFac | 1 | - | Snapshot stride when making these merger trees (should be 1). |
| NtreesPerFile | 1 | - | The number of TreeX group in this file. |
| NhalosPerFile | 1 | - | The total number of tree members (subhalos) in this file. Equals the sum of all elements of TreeNHalos. |
| NumberOfOutputFiles | 1 | - | Number of file chunks over which the merger tree is split. |
| 1 |
\(10^{10} M_\odot/h \) |
The dark matter particle mass used to make these merger trees. |
|
TreeX groups.
The datasets available for each one of them is provided below (click to open). Each entry refers to one subhalo, which can reside in any snapshot of the simulation.
| Datasets of TreeX groups ▾ |
|---|
| Dataset | Dim. | Description |
|---|---|---|
| SubhaloNumber | TreeNHalos[X] | The ID of this subhalo, unique within the full simulation for this snapshot. Indexes the Subfind group catalog at SnapNum. |
| Descendant | TreeNHalos[X] | The index of the subhalo's descendant within the merger tree, if any (-1 otherwise). Indexes this TreeX group. |
| FirstProgenitor | TreeNHalos[X] | The index of the subhalo's first progenitor within the merger tree, if any (-1 otherwise). The first progenitor is defined as the most massive one (-1 if none). Indexes this TreeX group. |
| NextProgenitor | TreeNHalos[X] | The index of the next subhalo from the same snapshot which shares the same descendant, if any (-1 if this is the last). Indexes this TreeX group. |
| FirstHaloInFOFGroup | TreeNHalos[X] | The index of the main subhalo (i.e. the most massive one) from the same FOF group. Indexes this TreeX group. |
| NextHaloInFOFGroup | TreeNHalos[X] | The index of the next subhalo from the same FOF group (-1 if this is the last). Indexes this TreeX group. |
| FileNr | TreeNHalos[X] | File number in which the subhalo is found. (Redundant, i.e. for a given [chunkNum] file, this array will be constant and equal to [chunkNum]) |
| SnapNum | TreeNHalos[X] | The snapshot in which this subhalo was found. |
| Most all original fields from the group catalogs are also available, reordered into the same order as the merger trees for convenience. See the group catalog description for their units, dimensions, and descriptions.
Fields: Group_M_Crit200, Group_M_Mean200, Group_M_TopHat200, SubhaloBHMass, SubhaloBHMdot, SubhaloCM, ... Note: Group_M_Crit200, Group_M_Mean200, and Group_M_Tophat200 are FOF group quantities, so that only the first subgroup from each FOF group will have a nonzero value. |
||
Merger tree description
The figure below shows a schematic of the structure of the merger trees. Walking back in time following along the main (most massive) progenitor branch consists of following the first progenitor links until they end (value equals -1). Similarly, walking forward in time along the descendants branch consists of following the descendant links until they end (value equals -1), which typically occurs at the last snapshot of the simulation (\(z = 5.5\)). The full progenitor history, and not just the main branch, requires following both the first and next progenitor links. In this way the user can identify all subhalos at a previous snapshot which have a common descendant.
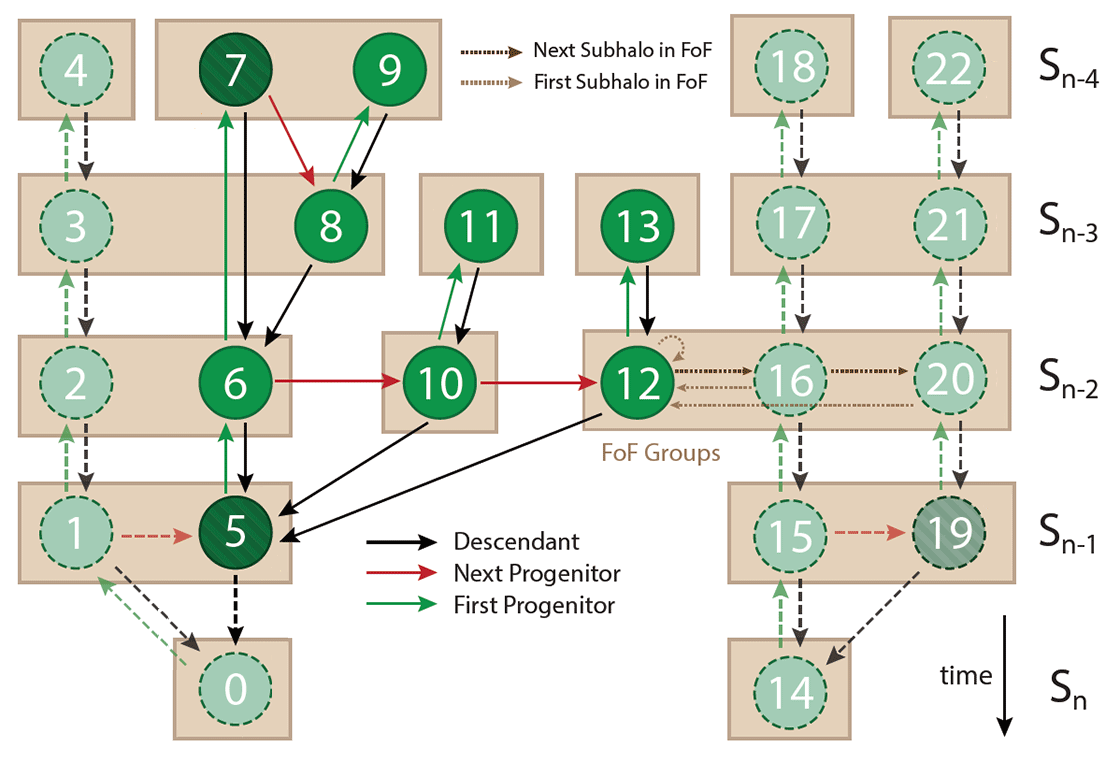
The number inside each circle from the figure is the unique ID (within the whole simulation) of the corresponding subhalo, which is assigned in a depth-first fashion. The ordering within a single tree in the LHaloTree is not guaranteed to follow this scheme.
Each subhalo spans a "subtree" consisting of the subhalo itself and all its progenitors. As an example, the subhalos belonging to the subtree of subhalo 5 are shown in darker green in the figure. Other subhalos not belonging to this subtree are shown in lighter green, and their links are indicated with dashed arrows.
The trees contain the links 'first subhalo in FoF group' (light brown dotted arrow) and 'next subhalo in FoF group' (dark brown dotted arrow), which connect subhalos that belong to the same FoF group. The FoF groups do not play a direct role in the construction of the merger tree.
The LHaloTree algorithm is virtually identical to that used for the Millennium simulation, constructing trees based on subhalos instead of main halos, described fully in the supplementary information of Springel et al. 2005. In short, to determine the appropriate descendant, the unique IDs that label each particle are tracked between outputs. For a given halo, the algorithm finds all halos in the subsequent output that contain some of its particles. These are then counted in a weighted fashion, giving higher weight to particles that are more tightly bound in the halo under consideration, and the one with the highest count is selected as the descendant. In this way, preference is given to tracking the fate of the inner parts of a structure, which may survive for a long time upon infall into a bigger halo, even though much of the mass in the outer parts can be quickly stripped. To allow for the possibility that halos may temporarily disappear for one snapshot, the process is repeated for Snapshot \(n\) to Snapshot \(n+2\). If either there is a descendant found in Snapshot \(n+2\) but none found in Snapshot \(n+1\), or, if the descendant in Snapshot \(n+1\) has several direct progenitors and the descendant in Snapshot \(n+2\) has only one, then a link is made that skips the intervening snapshot.
The LHaloTree merger tree is one large data structure split across several HDF5 files named trees_sf1_[NumLastSnapshot].[chunkNum].hdf5, where [chunkNum] goes from e.g. 0 to 1200 for Thesan-1.
Within each file there are a number of groups named TreeX, where X is an integer which simply increases from zero to the number of tree groups in that file chunk.
Note: Note that a given TreeX group may contain subhalos spanning different FoF groups as well as snapshots, and to efficiently locate a specific subhalo at a specific snapshot (e.g \(z=0\)) the offsets can be used.
The pair (SubhaloNumber,SnapNum) provides the indexing into the Subfind group catalog. The five other indices for each entry in a TreeX group (e.g. Descendant) index into that same group in the tree file.
